In my previous post I demonstrated array operations based on the new functionality I have implemented during my GSOC project. In this post I will discuss another feature that is essential for array calculations in a computer algebra system: The initialization of numeric arrays based on symbolic math expressions. After all, it is the symbolic stuff we are really interested in, isn’t it? 🙂 I will demonstrate how the automatic compilation and wrapping can be used to setup Numpy arrays with fast Hydrogen wave functions.
Again, it will be interesting to see how we compare with the array initialization features of numpy. One lesson I learned while preparing my previous post is that the timing results depend heavily on the compiler. To avoid the scaling issues we observed with the matrix-matrix product last time, I will do all examples on a computer with ifort 11.0. The Numpy installation on that computer is version 1.1.0. The warning applies again: I report the timing results I get on my computer, but this is not a rigorous benchmark. Also, I may not be using Numpy in the optimal way. If you know how I could make it run faster, I’d love hear about it.
Introductory example: linspace
First, lets try to create our own compiled linspace function that we can compare with numpy.linspace. The Numpy function creates an array of evenly spaced numbers, like this:
In [1]: import numpy as np In [2]: np.linspace(0, 1, 5) Out[2]: array([ 0. , 0.25, 0.5 , 0.75, 1. ])
The same functionality can be created by autowrapping a Sympy expression that maps the array index to an interval on the real axis:
In [3]: from sympy.utilities.autowrap import autowrap
In [4]: a, b = symbols('a b')
In [5]: i = Idx('i', m)
In [6]: linearmap = a + (i - 1)*(a - b)/(1 - m)
In [7]: x = IndexedBase('x')
In [8]: linspace = autowrap(Eq(x[i], linearmap))
In [9]: linspace(0, 1, 5)
Out[9]: [ 0. 0.25 0.5 0.75 1. ]
So, it worked. The array has been initialized according to the linear map defined by the Sympy expression at In[6]. In order to map the index to the interval correctly one must remember that the index of a Fortran array starts at 1, while C arrays start at 0. This must be be taken into account when the initialization expression is defined, so a slightly different expression must be used with the C backend.
Now, you may be wondering how I could know the order of the arguments in the wrapped linspace function, and why it was so conveniently compatible with the interface for numpy.linspace. The answer, is that unless autowrap gets the optional keyword argument ‘args’, the arguments are sorted according to their string representation. I carefully chose the symbols in the linear map so that my linspace would automatically get the same argument sequence as Numpy’s.
In [14]: %timeit np.linspace(0, 5, 100000) 1000 loops, best of 3: 916 µs per loop In [15]: %timeit linspace(0, 5, 100000) 1000 loops, best of 3: 381 µs per loop
Sweet! Evenly spaced numbers are of course very useful, but let us also try something a bit more juicy.
Universal hydrogen functions
One of the really useful concepts in Numpy is the “universal function” or ufunc. To put it short, ufuncs operate on arrays by applying a scalar function elementwise. It is actually more than that, as numpy ufuncs are required to support type casting, broadcasting and more, but we will ignore that and focus on the following quote from the Numpy docs:
That is, a ufunc is a “vectorized” wrapper for a function that takes a fixed number of scalar inputs and produces a fixed number of scalar outputs.
Can we make such functions with the autowrap framework? As you can probably guess, the answer is positive, so the question is rather: can we beat Numpy on speed? Let’s find out.
Very recently, Sympy got functionality to generate radial electronic wave functions for the Hydrogen atom. That is, quantum mechanical solutions to the attractive Coulomb potential. These are well known, and long since tabulated functions, that are routinely used in quantum mechanics. Wouldn’t it be nice to have the Hydrogen wave functions available as super fast ufunc-like binary functions?
The failsafe way to build a fast binary function that works element-wise on an array, is to construct a symbolic Lambda function that contains the initialization expression. The Lambda instance should then be attached to a regular Sympy Function. This is done with a call to implemented_function(‘f’, Lambda(…)) as in the following:
In [2]: from sympy.utilities.lambdify import implemented_function
In [3]: from sympy.physics.hydrogen import R_nl
In [4]: a, r = symbols('a r')
In [5]: psi_nl = implemented_function('psi_nl', Lambda([a, r], R_nl(1, 0, a, r)))
In [6]: psi_nl(a, r)
Out[6]: psi_nl(a, r)
In [7]: psi_nl._imp_(a, r)
Out[7]:
-r
____ --
/ 1 a
2* / -- *e
/ 3
\/ a
As you would guess from Out[6], psi_nl is just a regular Sympy Function, but with an additional attribute, _imp_, that works as displayed in In[7]. The implemented_function trick landed in Sympy just a few weeks ago. It was intended for numeric implementations, but in my autowrap3 branch, I (ab)use it to store a symbolic Lambda instead.
In [8]: x = IndexedBase('x')
In [9]: y = IndexedBase('y')
In [10]: i = Idx('i', m)
In [11]: hydrogen = autowrap(Eq(y[i], psi_nl(a, x[i])), tempdir='/tmp/hydro')
The argument ‘tempdir’ tells autowrap to compile the code in a specific directory, and leave the files intact when finished. Checking the Fortran source code reveals that the wave function is calculated like this:
do i = 1, m y(i) = 2*sqrt(a**(-3))*exp(-x(i)/a) end do
The implemented function has been printed as an inline expression, avoiding a costly function call in the loop. (If you wonder why I didn’t just print the expression directly, the explanation is here.) Now that we have the functions available, this is how they look:
The parameters and
are quantum numbers related to the electron’s energy and orbital angular momentum respectively.
The autowrapped Hydrogen functions can also be compared with numpy equivalents, as all waves are expressed in terms of functions that are universal in numpy. We use Sympy’s ‘lambdify’ to create a Python lambda function that calls the relevant ufuncs from numpy’s namespace:
In [31]: from sympy.utilities.lambdify import lambdify In [32]: psi_lambda = lambdify([a, r], R_nl(1, 0, a, r), 'numpy') In [33]: grid = np.linspace(0,3,100) In [34]: np.linalg.norm(psi_lambda(0.5, grid) - hydrogen(0.5, grid)) Out[34]: 2.21416777433e-15
But there are many solutions to the Hydrogen atom, so let’s study them a bit more systematically. The following is the output from a loop over the energy quantum number . I used only the s-wave functions
, corresponding to an electron that moves straight through the nucleus and out on the other side before turning back again to repeat the cycle. For each
, I calculated:
- %timeit for application of lambda function with Numpy ufuncs
- %timeit for application of the compiled and wrapped Sympy function
- the absolute difference between the solutions:
Here is the output:
n = 1 Numpy: 100000 loops, best of 3: 16.8 µs per loop Sympy: 1000000 loops, best of 3: 1.92 µs per loop difference: 1.07708980623e-14 n = 2 Numpy: 10000 loops, best of 3: 30 µs per loop Sympy: 1000000 loops, best of 3: 2 µs per loop difference: 5.13651621237e-15 n = 3 Numpy: 10000 loops, best of 3: 48.1 µs per loop Sympy: 100000 loops, best of 3: 3.37 µs per loop difference: 2.28896762655e-15 n = 4 Numpy: 10000 loops, best of 3: 70.9 µs per loop Sympy: 100000 loops, best of 3: 2.7 µs per loop difference: 6.18114500556e-15 n = 5 Numpy: 10000 loops, best of 3: 116 µs per loop Sympy: 100000 loops, best of 3: 4.61 µs per loop difference: 5.68967616077e-15 n = 6 Numpy: 10000 loops, best of 3: 142 µs per loop Sympy: 100000 loops, best of 3: 5.02 µs per loop difference: 1.21523884705e-14 n = 7 Numpy: 10000 loops, best of 3: 183 µs per loop Sympy: 100000 loops, best of 3: 5.95 µs per loop difference: 1.04406500806e-14
Awesome! For we beat Numpy by a factor of 30. 🙂 But the really interesting thing here is the scaling with expression complexity. Look at this:

Normalized execution time for Numpy and Sympy. As n increases, the complexity of the wave function expression increases, leading to the slowdown. The values of each curve are normalized against the corresponding n=1 calculation.
For higher the wave function expressions increase in complexity, and so does the execution time of both Numpy and Sympy. However, while the autowrapped Sympy expression needs 3 times longer for
than for
, the execution time for a lambda of native Numpy ufuncs increases tenfold!

Numpy execution time vs. Sympy. The mostly linear relation means that Numpy and Sympy have similar dependence on the complexity of the expression, although the scaling factors are quite different.
This last figure shows the linear relation between execution times of the compiled Sympy expressions and native Numpy lambda function. The plot supports the idea that the it is the inherent complexity of the mathematical expression that determines the execution time. By extrapolation, we can expect that for an expression that would take Sympy 9 times longer, Numpy would be hit by a factor of 100.
The main reason for the speedup, is that while Numpy calculates the wave function by applying a series of ufuncs, the Sympy way is to create a new “ufunc” and apply it once. It would be very cool to try this in an iterative solution scheme. With a super fast ufunc in a single python loop, one could do complex and cpu-intensive calculations with respectable performance from within isympy. That would be awesome!


That looks cool. When I used the R_nl() functions, I just used a lambda expression and it was quite slow. Your approach is way better.
I am really excited that the outcome of my gsoc project turned out to be this useful 🙂
Again an interesting post (and it looks nicer :)). Maybe I’ll try them later in your branch to see if it works this time, and if so, if I get similar results.
What did you use to create the plots? Did you automate it somehow, or did you have to enter the numbers manually? I would like to create a timing plot comparing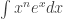 for
for  from 0 to 50, which is phenomenally faster in the new implementation), but I don’t want to manually grab 100
from 0 to 50, which is phenomenally faster in the new implementation), but I don’t want to manually grab 100
risch_integrate()tointegrate(), but I want to get a lot of points (say, compare%timeitnumbers.Yeah, it looks much nicer! Thanks a lot for teaching me 🙂
I created the plots with matplotlib. Ipythons %timeit uses the print statement, not return values, so I couldn’t automate it easily. For the number of datapoints I needed it was not a problem to enter the values manually, but for your plot I would suggest to use the timeit module directly, i.e. “import timeit”.
‘The main reason for the speedup, is that while Numpy calculates the wave function by applying a series of ufuncs, the Sympy way is to create a new “ufunc” and apply it once.”
It would be great to backport the Sympy behavior into Numpy (not as default because there is no point to “optimize” e=a+b+c+d this way if a,b,c,d and e are very small arrays).
I know about http://www.scipy.org/Weave but I’m not sure if it’s the “numpic” way to compile expressions like e=a+b+c+d into *one* ufunc….
That would be very cool indeed!
I think this is already done by numexpr:
http://code.google.com/p/numexpr/
Sorry, I get the same error as before (
ImportError: dynamic module does not define init function (initwrapper_module_0). I’m trying in the latestautowrap3branch (4496107) (funny how the first part of the SHA1 for your branches always seem to be pure numerical digits).Even without it working, one can see the cost of the faster code generated function, which is that it takes a minute or two initially to compile, even for a function as simple as the one in
[8]A suggestion: rename
quiet=Truetoverbose=False.For timeit, IPython’s
magic_timeit()has its own custom code to automatically determine the number of loops. So I think the solution is to just take the source for it and modify it to return a value instead of printing it. Or I noticed that Mateusz started a similar thing already in his polys11 branch, so maybe I will just work off of that.I agree that ‘verbose’ is a better choice than ‘quiet’ and Mateusz’ timeit looks very nice and useful. Thanks!
Maybe you have a problem with f2py. You can try on the command line:
It should then create the module “untitled.so”. I’m sorry I still haven’t found the time to implement Cython ndarray support.
There is of course an upfront time cost when you compile, but it should not take as long as a minute or two:
In [14]: from sympy.physics.hydrogen import R_nl In [15]: psi_nl = implemented_function('psi_nl', Lambda(r, R_nl(7, 0, 1, r))) In [16]: %timeit autowrap(Eq(y[i], psi_nl(x[i]))) 10 loops, best of 3: 2.14 s per loopf2py seems to work fine (
echo $?produces 0, and I get untitled.so).Well, I can’t test that exact one, because I don’t know where implemented_function comes from, but doing the one from the top of the original post, manually using f2py the command line with the bash time function, I get
real 0m19.028s
user 0m1.525s
sys 0m0.931s.
So maybe it’s just because I am running some other stuff at the same time 🙂
Sorry,
from sympy.utilities.lambdify import \ implemented_functionAre you able to “import untitled” ?
Nope. Importing untitled is where the ImportError comes in (as you might have guessed).
Cool! It would be very nice to have a fast tcc backend for autowrap. Also ctypes would be nice since it is in the standard libaries.
Hi,
I think your way of accelerating the calculation of symbolic expressions is exactly what I am looking for.
However, I do not seem to get the autowrap to work on my ubuntu maverick box.
I get the following error when trying to autowrap the linspace code:
In [9]: linspace = autowrap(Eq(x[i], linearmap))
—————————————————————————
CodeWrapError Traceback (most recent call last)
/home/bn/newpackages/python/sympy/ in ()
/home/bn/newpackages/python/sympy/sympy/utilities/autowrap.py in autowrap(expr, language, backend, tempdir, args, flags, verbose, helpers)
385 helps.append(Routine(name, expr, args))
386
–> 387 return code_wrapper.wrap_code(routine, helpers=helps)
388
389 def binary_function(symfunc, expr, **kwargs):
/home/bn/newpackages/python/sympy/sympy/utilities/autowrap.py in wrap_code(self, routine, helpers)
125 self._generate_code(routine, helpers)
126 self._prepare_files(routine)
–> 127 self._process_files(routine)
128 mod = __import__(self.module_name)
129 finally:
/home/bn/newpackages/python/sympy/sympy/utilities/autowrap.py in _process_files(self, routine)
149 if retcode:
150 raise CodeWrapError(
–> 151 “Error while executing command: %s” % ” “.join(command))
152
153 class DummyWrapper(CodeWrapper):
CodeWrapError: Error while executing command: f2py -m wrapper_module_0 -c wrapped_code_0.f90
Is there anything that can be repaired easily?
The same thing works on my mabook pro running 10.6.5
Ok, problem solved.
Needed to install the dev version of python to get the file Python.h
Good to hear that you solved it. For future reference: the best way to troubleshoot is to store the files with the tempdir keyword argument, and then run the command manually.
Pingback: Quick calculations with SymPy and Cython | studywolf